Using AI to Help Solve the Long COVID Puzzle
February 11, 2022
February 11, 2022
By Jonathan Woods
The condition has many names: post-COVID syndrome, long COVID, and post-acute sequelae of SARS-CoV-2. It’s COVID-19’s agonizing echo, and many have had the misfortune of experiencing it. More than half of those who’ve had COVID-19 have reported at least one lingering symptom three months after infection. According to the Public Health Agency of Canada, common long COVID symptoms include fatigue, shortness of breath, and memory problems, though “more than 100 symptoms or difficulties with everyday activities” have been reported. For some, the condition is debilitating and seemingly indefinite. [1]
Long COVID isn’t well-understood. “We don’t know what causes post COVID-19 condition,” reads the public health agency’s website. “There’s currently no single way to diagnose post COVID-19 condition,” and “[t]here’s currently no treatment.” [1]
Can AI help expedite the discovery of answers?
The Vector Institute’s Industry Innovation and AI Engineering teams worked with Roche Canada, Deloitte and TELUS to explore this question. Their project: applying natural language processing (NLP) techniques to social media posts made by people with long COVID to see if any patterns arise. This involved creating a machine learning pipeline and testing the capabilities of various NLP models on first-hand testimony from social media. The hope is that these patterns, if identified, could reveal clues about when and how frequently symptoms arise and where clusters of the condition occur. Any insights could be shared with clinicians to hone their research questions, identify trends early, or inform treatment strategies.
“Social media is often turned to by patients as an outlet to express their experience with illness,” reads Social Media Use in Healthcare, a research paper on how patients use social networks. Informational, emotional, and esteem support on social networks “commonly lead[s] to patient empowerment,” the paper continues. [2] This encourages sharing, and can make social media a rich resource for researchers ― but only if they can reliably filter the ocean of posts made every day and pinpoint those with relevant language.
Even for advanced machine learning models, this is a challenge.
Elham Dolatabadi, Applied Machine Learning Scientist at the Vector Institute and the project’s technical lead, says, “Extracting medical entities from social media is challenging because of the unstructured nature of the content, which is often noisy, informal, and short. That is not to mention that the complexity of medical terms sometimes results in misspellings.” User generated posts are anything but uniform, and their brevity and lack of structure (which includes the use of slang and differences in tone) make identifying, extracting, and classifying first-hand accounts of long COVID difficult.
To address these challenges, the team developed a customized machine learning (ML) pipeline specifically designed to find, order, and display long COVID terms that would otherwise be buried in a mountain of social media posts. The pipeline organizes the process from end to end, bringing the collection and filtering of data, training of various models to extract and classify key terms from posts, and visualization of results on dashboards into one process tailor-made for the task.
Step one in the development of the pipeline was creating datasets made up of posts from Twitter and Reddit. Using Twitter’s application programming interface (API), the team searched for tweets that included relevant hashtags ― like #longcovid, #postcovidsyndrome, and #covidlonghauler ― along with similar, non-hashtagged terms. Once collected, they ran the data through a long-hauler filter designed to identify first-person accounts. This process also involved removing posts that include news headlines or proprietary product names. The remaining tweets were de-identified, with all personal information about the poster removed (though timestamps, geographical information, and general user profile descriptions remained).
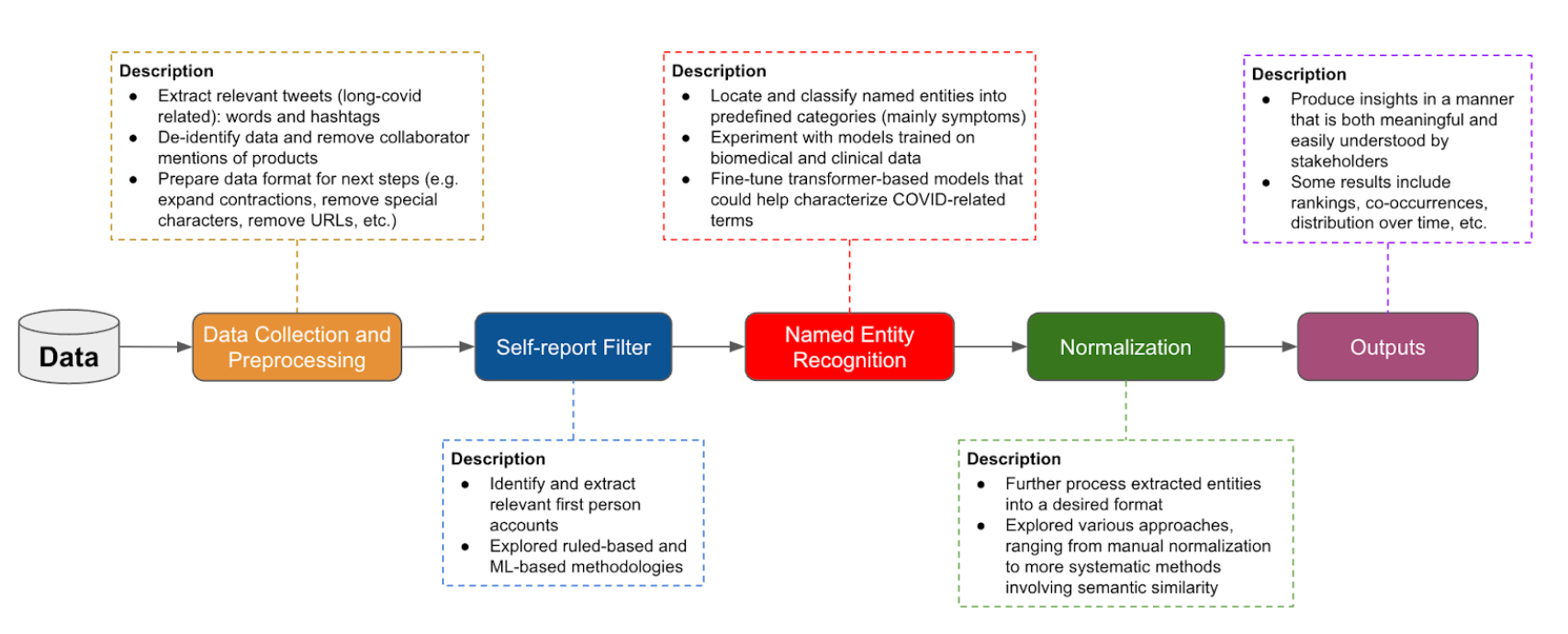
Figure 1. The team built a customized ML pipeline for collecting, filtering, analyzing, and visualizing long COVID information gleaned from social media posts.
With datasets in hand, the team tested an ensemble of specialized models trained in an NLP technique called named entity recognition (NER). NER identifies specific entities (like people, places, and objects) in text and then classifies them into predefined categories. These experiments were designed to see if models, trained specifically to extract medical terms, could identify language denoting a long COVID symptom, test, or treatment, and then accurately classify it as such. Figure 2 illustrates how this works on two sample tweets.
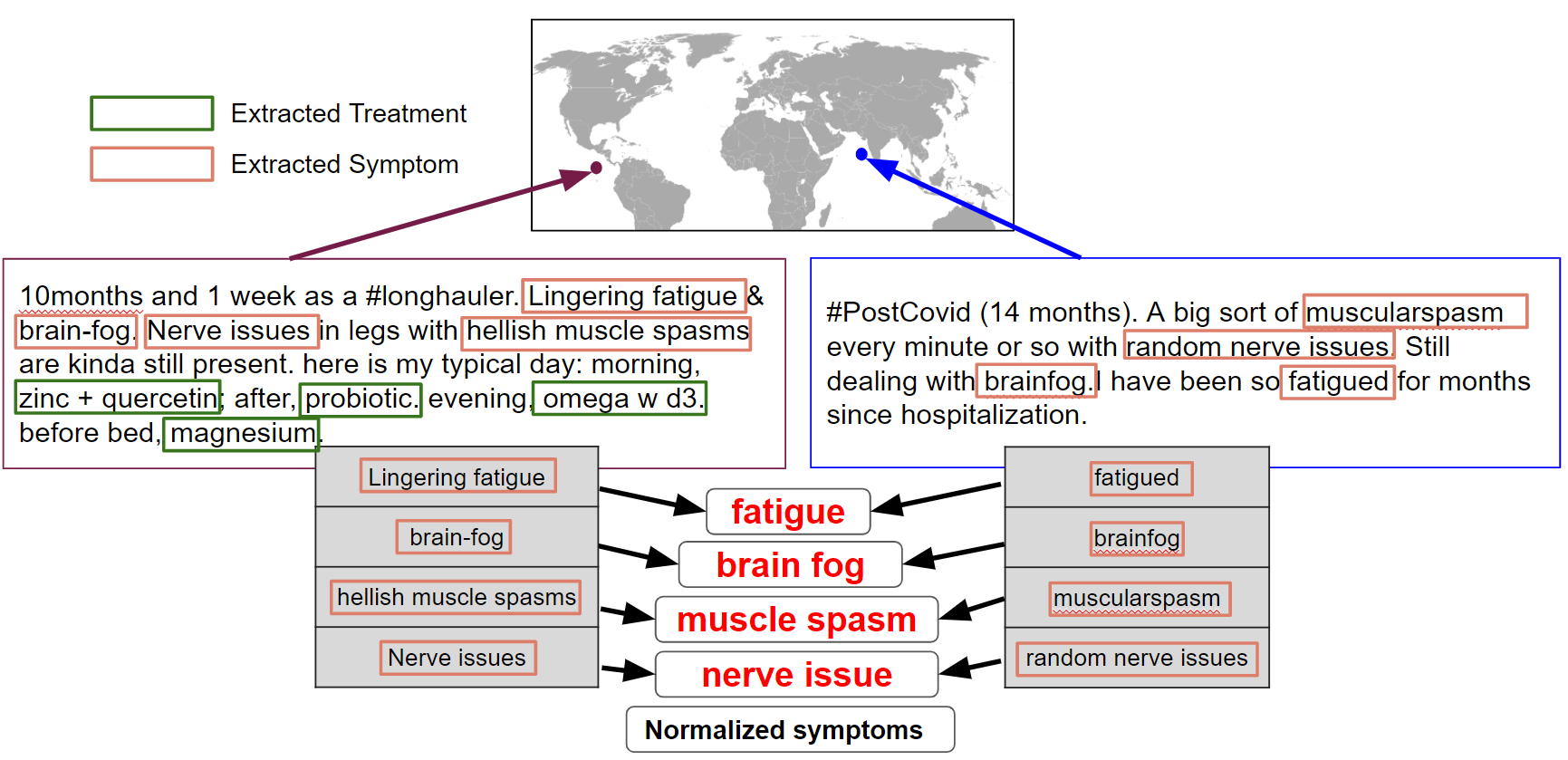
Figure 2. Among the models the team explored was Stanza, an NER model created by Stanford. Stanza was trained on a corpus of biomedical and clinical text and is used to identify symptoms or treatments in social media posts, and extract, classify, and normalize them appropriately. Note that the examples are synthetically created.
One of the models the team tested was UmlsBERT, a model already trained on a large body of clinical Metathesaurus (UMLS Metathesaurus). Part of the experiment was fine-tuning this model on clinical related dataset provided by National Center for Biomedical Computing (NCBC) known as i2b2 and data augmenting to enable more granular entity extraction. Using UMLS’ MetaMapLite and AMIA Task3 dataset the team improved the entity extraction to be able to pick out even colloquial and informal terms, like ‘brain fog’ and ‘crushing fatigue.’
After conducting a series of experiments, preliminary results showed that patterns related to symptom frequencies, co-occurrence, and distribution over time could be successfully detected and visualized. Researchers can indeed find signals in the social media noise.
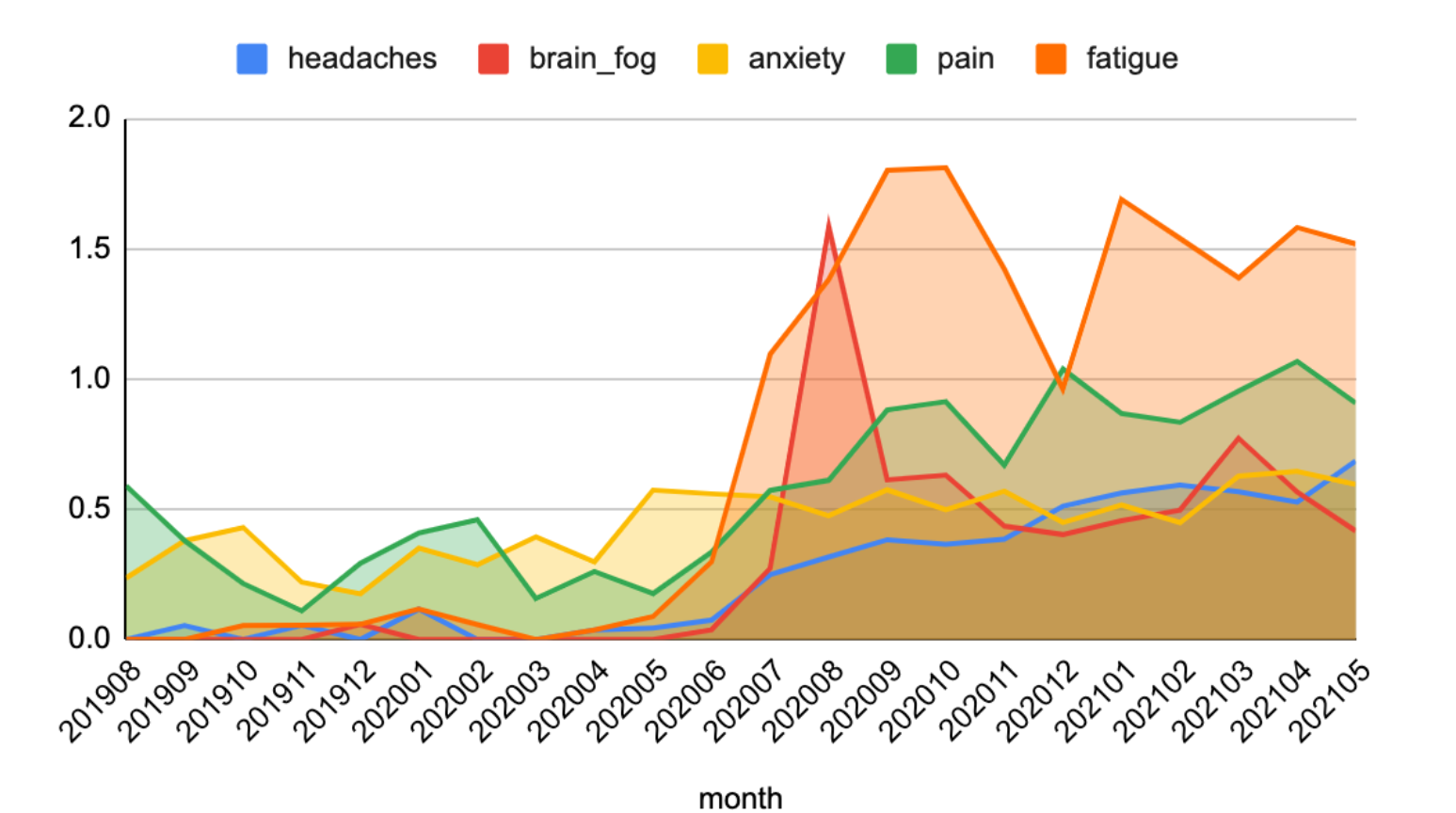
Figure 3. The occurrence frequency of top five (5) symptoms over time using the Stanza model and Twitter data with a specific COVID filter and normalization.
With confirmation of the pipeline’s effectiveness for COVID-related entity extraction from social media, the next step is to work with researchers and clinicians to interpret these patterns and determine what promising research directions they can reveal.
“We can use this approach to give clinicians more insight and provide early signals that can help them plan their clinical studies, treatments, therapeutics, and so on,” says Vector project manager Sedef Akinli Kocak. “This is a great example of collaboration between researchers, subject matter experts, and the companies that would like to contribute to combatting COVID 19.”
The participating companies, medical subject matter experts, and the contributions of PhD candidates from various Canadian institutions deserve special credit. Together, over the course of the project, Roche Canada, Deloitte, TELUS and PhD researchers provided clinical and machine learning expertise that enabled the pipeline to function. Key contributions by these Vector sponsor companies and PhD researchers include original project ideation; review of clinical literature on COVID long haulers; data collection, cleaning, and annotation; filter implementation; normalization; NER modelling; visualization engineering; and interpretation of results.
The value of these contributions will extend beyond this pandemic. Researchers now have a tool and process to make social media a key resource for understanding other population-level health events, such as new emerging infectious diseases, rare diseases, or the effects of booster shots on infection in various areas at various times.
For today though, the focus remains on those experiencing long COVID. The hope is that this new ability can bring researchers and clinicians one step closer to solving the mysteries of long COVID and play a role in alleviating the suffering of those feeling its effects.
A technical paper related to the project has been accepted for presentation at the 6th International Workshop on Health Intelligence at the AAAI 2022 Conference. A full project report with outcomes will be released in early 2022. For now, the project remains in progress.
References:
[2] Smailhodzic, Edin, et al. “Social media use in healthcare: a systematic review of effects on patients and on their relationship with healthcare professionals.” BMC health services research 16.1 (2016): 1-14. LINK: https://


